Hannon Group
Small RNAs and mammalian genomics

Introduction
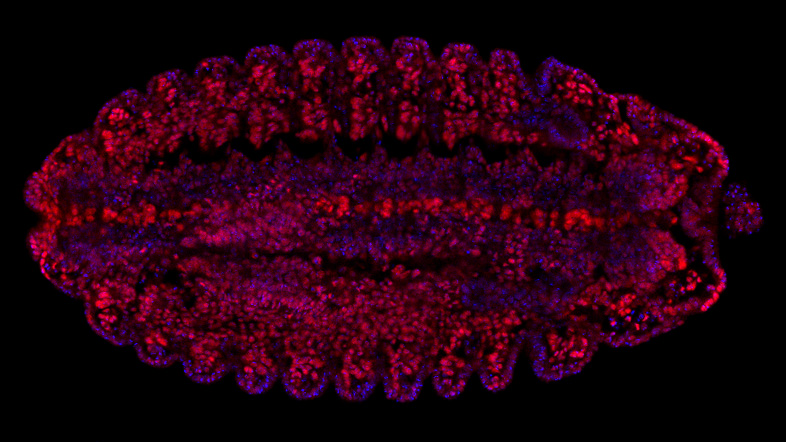
The Hannon Group has a long-standing interest in small RNAs, having defined many key components of the RNAi and microRNA pathways (Hammond et al., 2000, Bernstein et al., 2001, Hammon et al., 2001 and Liu et al., 2004). This led to a particular interest in a germline-specific class of small RNAs, called PIWI-interacting RNAs (piRNAs), which form part of a sophisticated innate immune system that essentially distinguish genes (self) from transposons (nonself) and selectively silences potentially active mobile elements (Girard et al., 2006, Brennecke et al., 2007, 2008, Malone et al., 2009).
Over the past decade, we have uncovered the components of the piRNA pathway and continue to uncover exciting new discoveries of the host-parasite arms race (Czech et al. 2013, Muerdter et al 2013, Czech et al., 2018, Munafo et al., 2019, Fabry et al., 2019, Kneuss et al., 2019, Eastwood et al., 2021, Munafo et al., 2021, Fabry et al., 2021, van Lopik et al., 2023). Our interest in small RNAs and RNAi motivated the development of short hairpin (shRNAs) as powerful tools for mammalian genetics (Paddison et al., 2002). These advances along with our development of in-situ oligonucleotide synthesis on microarrays as a way to generate complex oligonucleotide libraries (Cleary et al., 2004), allowed the development of genome-wide shRNA libraries for several animal models, which are now publicly available and widely used (Silva et al., 2008). These oligonucleotide technologies also facilitated the development of exome capture (Hodges et al., 2007), and related genome partitioning strategies, now widely utilised in cancer genomics, as well as the development of CRISPR libraires.
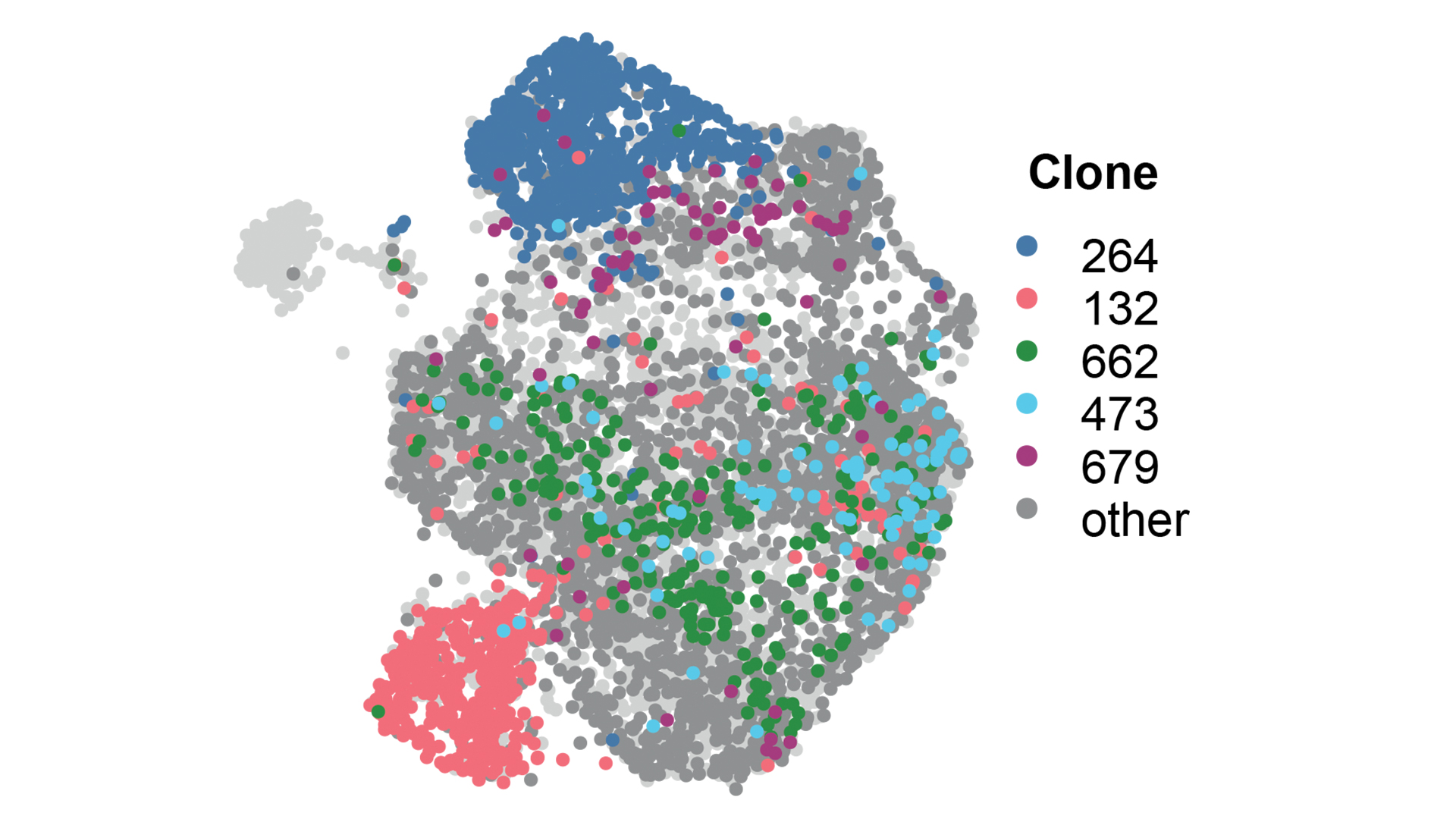
More recently, the group has pioneered dual guide CRISPR screening approaches, defining rules that make potent guide pairs to maximise loss of function outcomes (Erard et al., 2017).
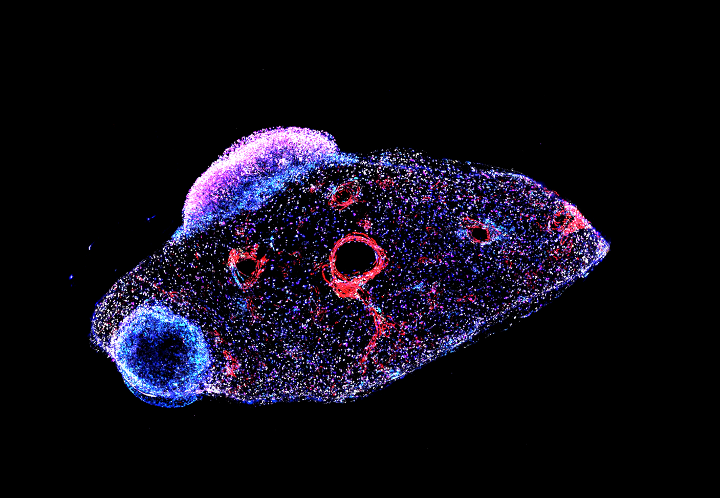
Current research within the group involves a continued interest in small RNAs and chromatin biology, understanding the biology of early breast cancer, and developing tools and techniques to track tumour heterogeneity.

Professor Greg Hannon
Director,
Senior Group Leader
Research themes
Related News
See all news-

Prof Greg Hannon shortlisted for prestigious Cancer Grand Challenge
24th September 2025
The shortlist of 12 multidisciplinary, global teams is now competing for up to £20m each, with the aim of delivering breakthroughs that no single researcher, lab, institute or country could achieve alone.
Find out more -
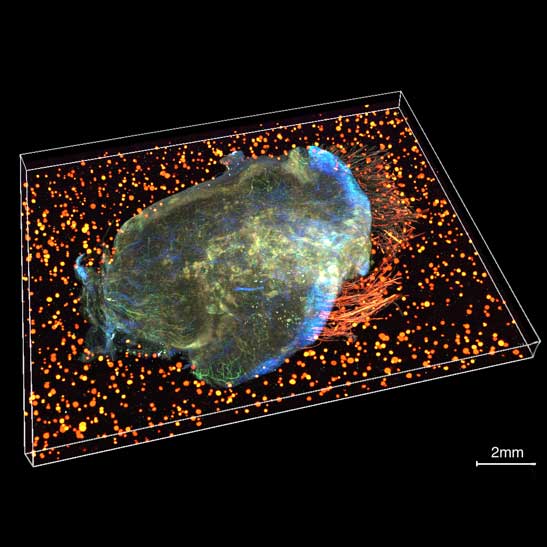
Scientists awarded £5m to advance groundbreaking tumour mapping technology
13th September 2024
The IMAXT team have been awarded £5.2 million to establish the Spatial Profiling and Annotation Centre of Excellence (SPACE) to open up access to their groundbreaking cancer mapping technology to the research field.
Find out more -
$6.5 million awarded to develop precision breast cancer research in Cambridge
30th October 2023
The project aims to help doctors predict the best treatment for patients with high-risk breast cancers.
Find out more
Publications
-
A dual histone code specifies the binding of heterochromatin protein Rhino to a subset of piRNA source loci
E-pub date: 13 Jan 2024
-
Ovo is a master regulator of the piRNA pathway in animal ovarian germ cells
E-pub date: 27 Apr 2024
-
Spatially tuneable multi-omics sequencing using light-driven combinatorial barcoding of molecules in tissues
E-pub date: 25 May 2024
-
Unistrand piRNA clusters are an evolutionarily conserved mechanism to suppress endogenous retroviruses across the Drosophila genus
E-pub date: 13 Nov 2023
Laboratory Efficiency Assessment Framework (LEAF)
The Hannon Group contributed to the Institute’s LEAF Silver accreditation, see the Sustainability webpage for more information.