Creixell Group
Cancer signalling and therapeutics

Research summary
In our laboratory, we develop and integrate computational and experimental technologies ranging from machine learning, drug design and functional protein biochemistry with the long-term-goal to make an impact in our understanding and treatment of cancer and drug resistance.
Introduction
Our lab integrates machine learning and high-throughput biochemistry to study how proteins selectively recognize their substrates, how this process is perturbed in cancer and how it can be hijacked to find highly selective and mutant-specific drugs to overcome drug resistance.
Targeted therapies have significantly improved outcomes for patients and shifted the clinical and biological goal towards targeting evolutionary trajectories and overcoming resistance. To overcome these challenges, it is critical to repurpose existing cancer drugs and design news ones with higher selectivity, lower toxicity, and less prone to resistance.
In our lab we combine and develop technology ranging from peptide display, deep sequencing, machine learning, drug design and functional protein biochemistry with the long-term goal to make an impact in our understanding and treatment of cancer and drug resistance. Our previous studies have taught general principles in cellular signalling specificity, which we are now using to investigate unexplored cancer signalling, molecular recognition and epistasis, novel therapeutics and predict and overcome drug resistance.

Dr Pau Creixell
Junior Group Leader
Group Members
-

Carol Mendonca
Senior Scientific Associate
-

Mihkel Ord
Postdoctoral Researcher
-

Gerard Duart
Marie Curie Fellow
-

Mingxuan Jiang
Postgraduate Student
-

Nuo Cheng
Postgraduate Student
-

Luis Bermudez-Guzman
Postgraduate Student
-

Amelia Barclay
Postgraduate Student
-

Jun Jie Peng
Postgraduate Student
-

Ocean Tsang
Postgraduate Student
-

Sian Evans
Postgraduate Student
-

Sydney Porto
Research Assistant
-

Mohan Sun
Research Assistant
Related News
See all news-
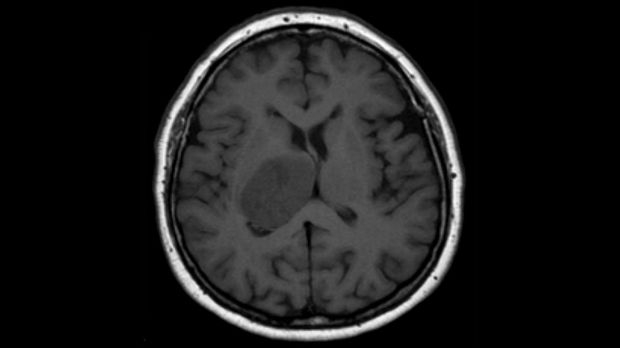
Quest for Cures: Glioblastoma research receives £1.5 million from The Brain Tumour Charity
31st July 2024
The Creixell Group has been awarded a £1.5 million ‘Quest for Cures’ grant from The Brain Tumour Charity for research aimed at improving treatments for glioblastoma.
Find out more -

Dr Pau Creixell joins Institute as new Group Leader
28th September 2020
Dr Pau Creixell has joined the Institute as a new Junior Group Leader, in partnership with the CRUK Cambridge Centre and Children’s Brain Tumour Centre of Excellence.
Find out more
Laboratory Efficiency Assessment Framework (LEAF)
The Creixell Group contributed to the Institute’s LEAF Silver accreditation, see the Sustainability webpage for more information.