Proteomics

Expert services for protein identification, quantification, and post–translational modification profiling using next–generation mass spectrometry platforms.
Our team collaborates with research groups to provide access to specialist mass spectrometry and proteomics equipment and expertise. We support protein identification, characterisation and quantification, post-translational modification profiling, and targeted analyses using state-of-the-art mass spectrometry platforms.
We can help design experimental strategies, implement and validate previously developed proteomic workflows to profile proteins from diverse biological samples, as well as develop entirely new, bespoke methods and assays when required. We strive to increase the scope, sensitivity, and throughput of proteomics technologies and their application to cancer-related questions.
The Core focuses and specialises in using stable isotopic labelling strategies (Tandem Mass Tags) as well as label-free methods (DDA and DIA) for the relative quantification of protein expression levels and enrichment within protein complexes. We also develop and run PRM (parallel reaction monitoring) assays for targeted quantification of defined panels of proteins.

Dr Clive D’Santos
Head of Proteomics
Proteomics help
We are open to collaborations with researchers from outside of the Institute, and would welcome the opportunity to discuss your project.
Please get in touch via our contact form on the Working with the Core Facilities page to explore the support we can provide.
Areas of interest
Equipment and analysis
-

Orbitrap Mass Spectrometers
We have three state-of-the-art mass spectrometers: Astral-Zoom, Ascend and Fusion Lumos, all of which are configured with a Vanquish Neo nano-HPLC system.
-

Off-line Separation Platforms
We operate two offline Dionex Ultimate 3000 capHPLC systems to provide multidimensional protein- and peptide-level separation, complementing our mass spectrometry workflows.
-

Relative quantification
Our quantification services include label-free methods (DDA and DIA), label-based approaches (SILAC and TMT), and targeted proteomics using PRM.
-

Data analysis
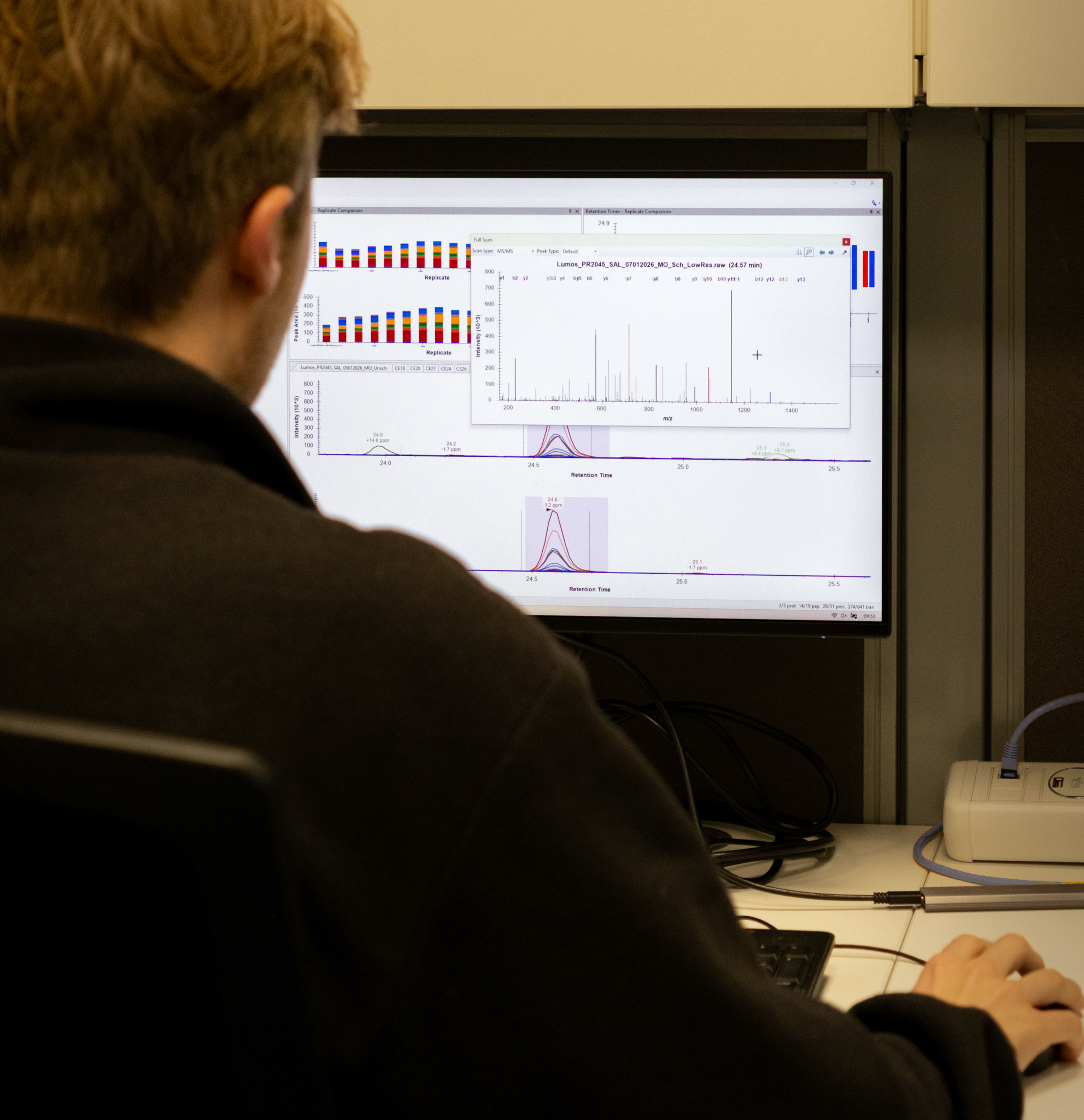
Data analysis is performed by Core Facility staff using the latest software, including DIANN, Proteome Discoverer, and Skyline. Additional statistical analysis can be provided by the Bioinformatics Core Facility upon request.
Related News
See all news-
New immune pathway offers treatment hope for childhood brain tumours
3rd February 2026
A newly discovered immune pathway could lead to gentler treatments for multiple childhood brain cancers, according to new research from our Gilbertson Group published today in Nature Genetics.
Find out more -

Institute scientists uncover molecular switch that drives pancreatic cancer progression
30th October 2025
New research from our Carroll Group has identified a molecular mechanism that helps explain how pancreatic ductal adenocarcinoma progresses, offering a potential path toward more targeted treatments.
Find out more -
Proteomics Facility awarded £1.3 million to boost capabilities
30th January 2024
Dr Clive D’Santos awarded £1.3 million to acquire a state-of-the-art mass spectrometer.
Find out more
Laboratory Efficiency Assessment Framework (LEAF)
Proteomics contributed to the Institute’s LEAF Silver accreditation, see the Sustainability webpage for more information.