Gilbertson Group
Cellular and molecular origins of cancer

Research summary
Our research focuses on understanding the origins of cancer, both in children and adults. Our studies of childhood brain tumours seek to understand how these tumours are born in the developing brain, and therefore how we can better classify and treat these diseases; generating cures that have minimal long-term side effects. Since development continues from birth throughout aging, we are applying similar principals to understand the origins and treatment of cancers in adults.
Introduction
The principal aim of my research is to improve the accuracy of brain tumour classification and treatment, avoiding long-term side effects for those who can be cured, whilst providing new therapeutic targets to guide future treatment. With a particular focus on children’s brain tumours. We are working to understand the cellular and molecular origins of cancers and the pathways that drive them. We hope to achieve this goal by:
- Conducting extensive genomic analyses of brain tumours to identify cancer-causing genetic abnormalities
- Understanding the impact of signalling pathways on normal stem cell biology and tissue development
- Integrating studies of genetic and cell signal alterations in cancers with studies of normal progenitor cells to determine the cellular and molecular origins of tumours
- Translating knowledge of tumour biology into effective new cancer cures through pre-clinical and early clinical trials of molecular targeted therapies

Professor Richard J Gilbertson FRS, FMedSci, FRCP, EMBO
Senior Group Leader
Research areas
Related News
See all news-
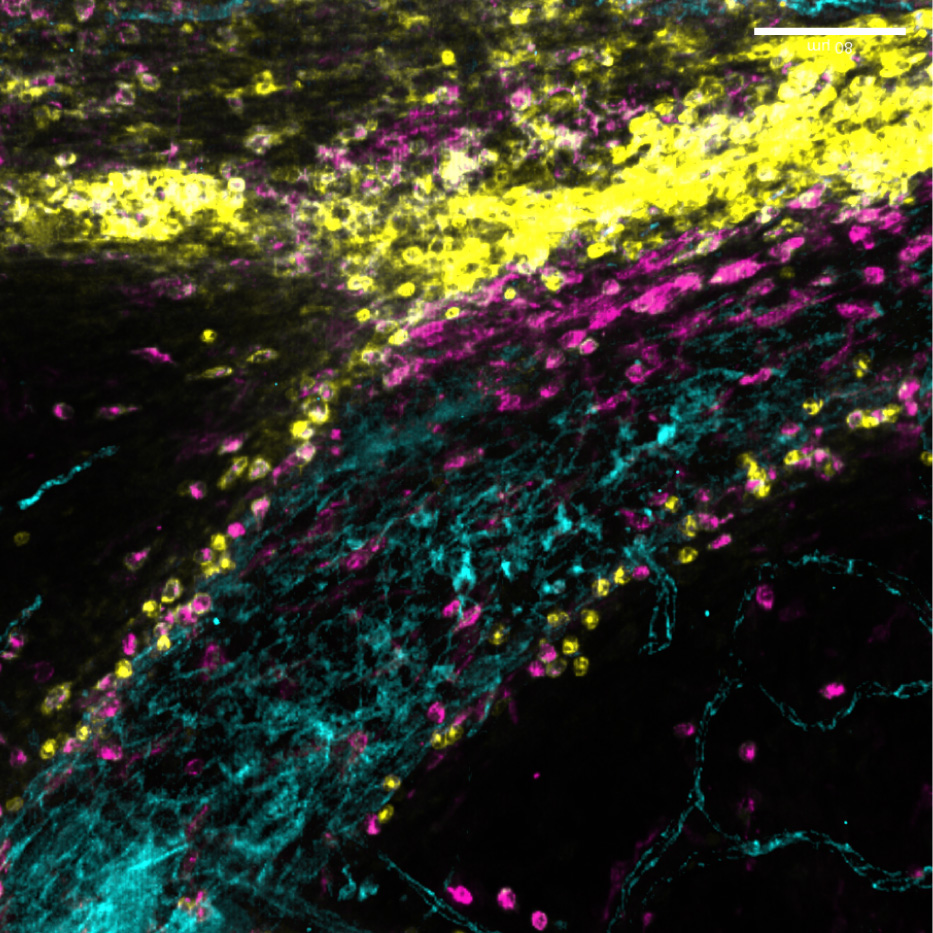
New immune pathway offers treatment hope for childhood brain tumours
3rd February 2026
A newly discovered immune pathway could lead to gentler treatments for multiple childhood brain cancers, according to new research from our Gilbertson Group published today in Nature Genetics.
Find out more -

Study looks at radiotherapy to treat and diagnose children’s brain tumours and avoid invasive surgery
3rd April 2025
A world first Cancer Research UK study is looking at radiotherapy to treat and diagnose children’s brain tumours and avoid invasive surgery.
Find out more -

Cambridge children’s cancer scientists to share £3M boost
30th September 2024
Cambridge researchers are set to receive a major cash injection from Cancer Research UK to help develop the next generation of treatments for children with brain tumours.
Find out more
Publications
-
Cancers make their own luck: theories of cancer origins.
E-pub date: 1 Oct 2023
-
The NALCN channel regulates metastasis and nonmalignant cell dissemination.
E-pub date: 1 Dec 2022
-
ZFTA Translocations Constitute Ependymoma Chromatin Remodeling and Transcription Factors.
E-pub date: 1 Sep 2021
-
DDX3X Suppresses the Susceptibility of Hindbrain Lineages to Medulloblastoma.
E-pub date: 24 Aug 2020
Laboratory Efficiency Assessment Framework (LEAF)
The Gilbertson Group contributed to the Institute’s LEAF Silver accreditation, see the Sustainability webpage for more information.