Balasubramanian Group
Chemical biology
Research summary
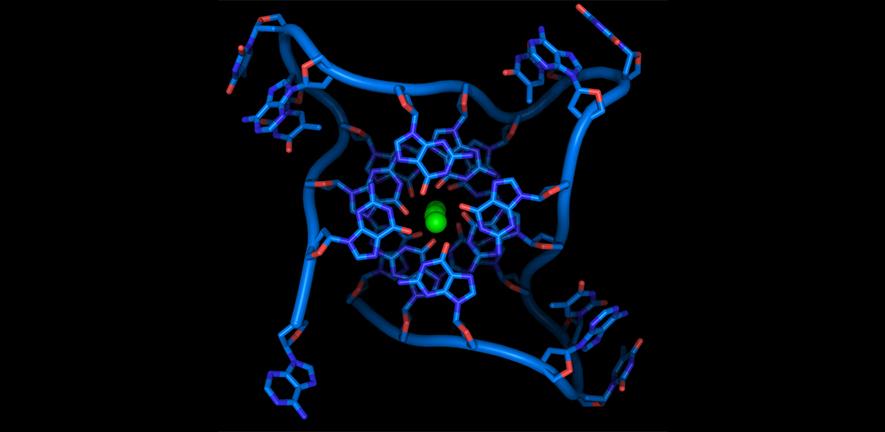
We investigate special DNA structures called quadruplexes that are involved in the expression of cancer genes. Using chemical biology methods, we have invented small drug-like molecules that bind to these structures, stabilising them. This can switch off cancer genes and thus stop cancers growing. These drug-like molecules may work on many different cancer genes making them potentially much less toxic for non-cancer cells than conventional drugs.
Introduction
Recent advances in understanding nucleic acid function have shown that alternative secondary structures and the chemical modification of nucleotide bases have key regulatory roles in diverse cellular processes, from transcription and translation to cell division and genome stability.
Genetic information is carried not only by the sequence of nucleic acids, but also their secondary structures and chemical modifications. For example, guanine-rich sequences can form stable four‑stranded structures called G-quadruplexes (G4s), while certain cytosine bases in DNA can become methylated. We hypothesise that such alternative structures, or chemical modifications, have critical functions in normal cells and cancer. By identifying where base modifications and G4 structures are located in the cancer cell genome, and through the application of synthetic small molecules that selectively target G4 structures, we aim to understand the oncogenic process and develop novel approaches for potential use in treatment and diagnosis of cancer. We are also exploring new strategies to target the DNA binding activity of FOXM1, a key cancer-related transcription factor involved in cell cycle control.

Professor Sir Shankar Balasubramanian
Senior Group Leader
Research areas
Related News
See all news-

Prof Sir Shankar Balasubramanian Awarded Prestigious Royal Society of Chemistry Prize
25th June 2025
Professor Sir Shankar Balasubramanian has received the Royal Society of Chemistry Khorana Prize, awarded for outstanding contributions through work at the chemistry and life science interface.
Read more -

Sir Shankar Balasubramanian elected Fellow of the AACR Academy
10th March 2025
Sir Shankar has been recognised for his pioneering research in nucleic acid chemistry and his key role in the development of next-generation sequencing technology.
Find out more -

Isabel Esain Garcia awarded PhD Thesis Prize
3rd December 2024
The Prize is awarded annually to a student who has undertaken an outstanding research project to the highest standards during the course of their PhD study at the Cancer Research UK Cambridge Institute.
Find out more
Publications
-
Improved simultaneous mapping of epigenetic features and 3D chromatin structure via ViCAR.
E-pub date: 27 Nov 2024
-
Spatially tuneable multi-omics sequencing using light-driven combinatorial barcoding of molecules in tissues
E-pub date: 25 May 2024
-
An Upstream G-Quadruplex DNA Structure Can Stimulate Gene Transcription.
Balasubramanian Group
E-pub date: 15 Mar 2024
-
G-quadruplex DNA structure is a positive regulator of MYC transcription.
E-pub date: 13 Feb 2024
Laboratory Efficiency Assessment Framework (LEAF)
The Balasubramanian Group contributed to the Institute’s LEAF Silver accreditation, see the Sustainability webpage for more information.